Antibiotic resistance: from soil to food chain
By Rose Edwin, Ph.D. student, Teagasc
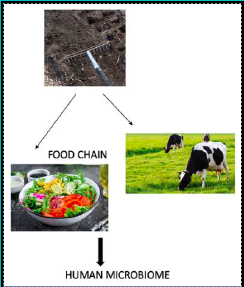
Fig 1. Transmission route of antibiotic resistance genes from soil to human microbiome
Antibiotic resistance has been recognised as a serious threat to human and animal health. The dispersal of antibiotics into the soil through anthropogenic activities has led to the selection, proliferation, and dissemination of antimicrobial resistance genes (ARGs) in the environment. There is a growing body of evidence that the ARGs in the soil can potentially enter the food chain and subsequently integrate into gut microbes. However, the sources of antimicrobial resistance in soil and its transmission into plants remain unclear.
Approximately 70% of soil microbes are uncharacterised; unraveling the complexity of soil microbiomes is our first challenge. As part of my work at VistaMilk, I will use state-of-the-art techniques including metagenomics and machine learning to unravel the DNA profile from 136 soil samples across Ireland. This will enable us to identify the unknown fraction of microorganisms present in the soil. Metagenomic sequencing also facilitates the characterisation of the genetic profile of the microbiome, which can be further examined for genes responsible for ARG. This will help us identify the taxonomy of Irish soil microbes and to analyse the prevalence of antimicrobial resistance genes across the country.
The project, upon completion, will enhance our knowledge of soil microbes in the food chain along with producing a reference soil microbiome database that will serve as an essential resource for soil researchers globally.
For more information please contact; N. Rose Edwin NiranjanaRose.Edwin@teagasc.ie

