Imagine a world where bacteria possess computing abilities akin to artificial neural networks, driving their responses to changing environments. This is not science fiction; it’s a fascinating reality researchers have been digging for years. Although we know that there is immense computational power inside a microscopic cell, harnessing it has been an unsolved problem. However, our latest research has unlocked the key to harnessing this incredible potential, revealing a method to convert the intricate Gene Regulatory Network into a Gene Regulatory Neural Network (GRNN, a term we introduce), paving the way to Living AI.
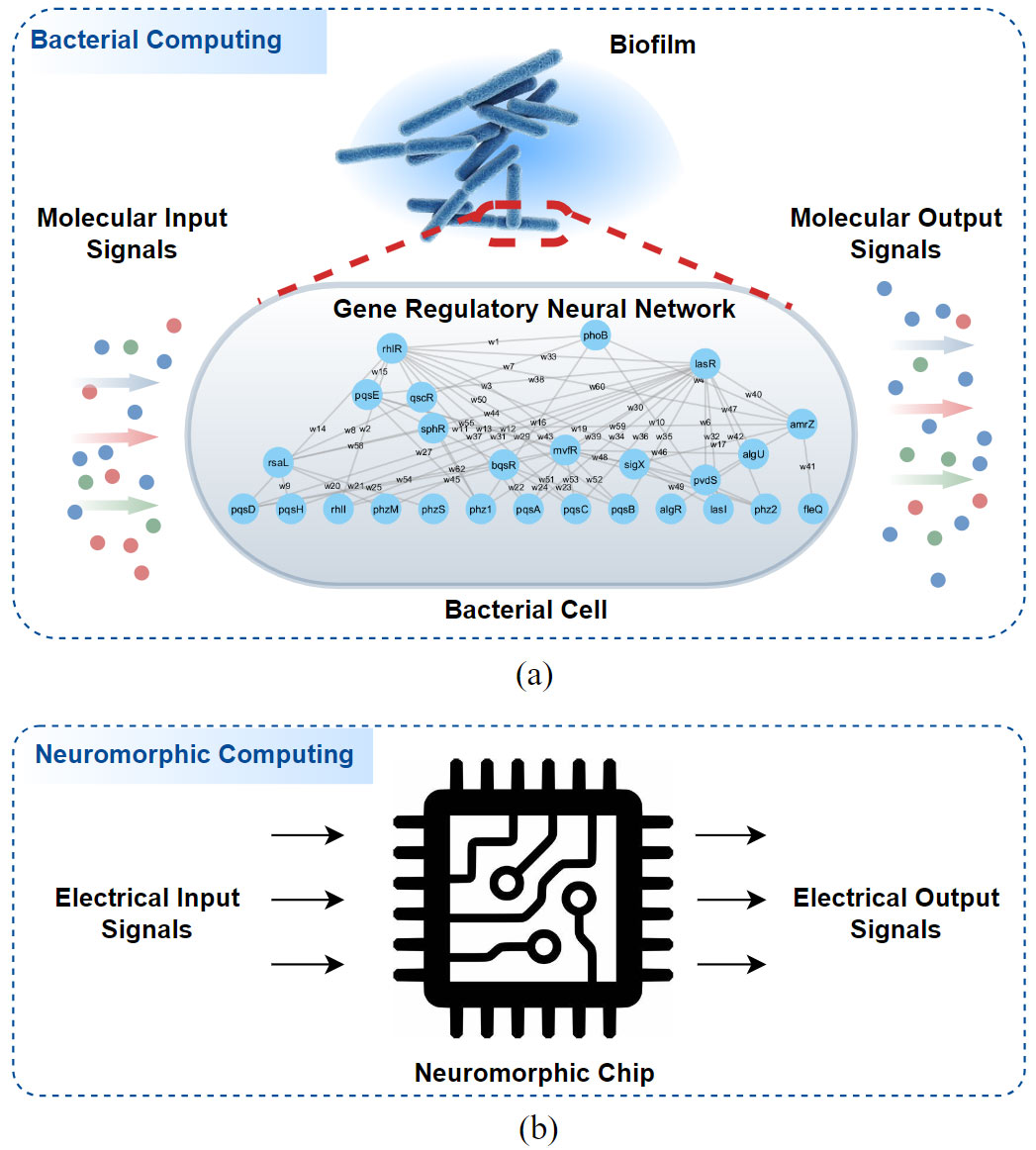
Fig: Illustration of the comparison between two computing mechanisms where a) is the introduced bacterial computing paradigm with Gene Regulatory Neural networks, and b) is the neuromorphic computing paradigm.
In the realm of computing, neural networks have taken center stage due to their remarkable capabilities in tasks like image recognition and natural language processing. These networks, inspired by the human brain, process information in layers of interconnected ‘neurons,’ making them a potent tool for complex tasks. While traditionally a soft concept, researchers have recently pioneered hardware-based neural networks known as Neuromorphic computing, which is far more powerful than conventional computing methods. This innovation transcends the limitations of conventional computing methods, mimicking the brain’s architecture to achieve unprecedented computational power and efficiency. Interestingly, our latest study found resemblance between Neuromorphic computing and bacterial decision-making process, as bacterial cells naturally contain a neural network-like arrangement of biological components.
In this study, we introduced a novel framework to unveil the intrinsic neural network-like computational structure within bacterial cells and tested it on the widely recognized Escherichia Coli (E. Coli) bacteria. Our exploration revealed a GRNN comprising approximately 5000 nodes, a remarkable complexity considering the microscale physical dimensions of around 2 micrometers.
However, bacteria rarely exist in isolation; they thrive in complex ecosystems such as biofilms. Bacteria within biofilms communicate using a sophisticated molecule-based signaling system, which operates constantly and is vital for their existence. We further proved that this constant communication directly influences the computational properties of individual cells, shaping their responses to stimuli.
Moreover, biofilms consist of a protective mechanism called extracellular polymeric substance (EPS); a thick slime acts as a barrier, minimizing the penetration of harmful substances. While providing safety, EPS also creates an environment with limited nutrient flow to cells at the core of the biofilm. This altered nutrient distribution triggers changes in cell-cell communication patterns that ripple through the biofilm, impacting its computing properties. As a result, we came up with an intriguing finding where a biofilm consists of a multi-layered computing architecture housing diverse computing capabilities. This intricate interplay between bacterial communication, protective mechanisms, and nutrient distribution creates a fascinating tapestry of computation within biofilms, offering insights into the complex yet effective bio-computing paradigm.
Our finding holds immense social implications that could reshape the bio-computing landscape in the future. Picture a world where computers not only process data but also learn and adapt like living organisms, and a medical diagnostic powered by biocompatible living AI, where bacteria do computing inside the body that assists in early disease detection by analyzing complex patterns in medical data. In addition, environmental monitoring could reach new heights as bacterial neural networks help detect pollution and respond to changing conditions in real-time at the site. This could lead to more effective, sustainable, and energy-efficient computing solutions for plethora of applications.

